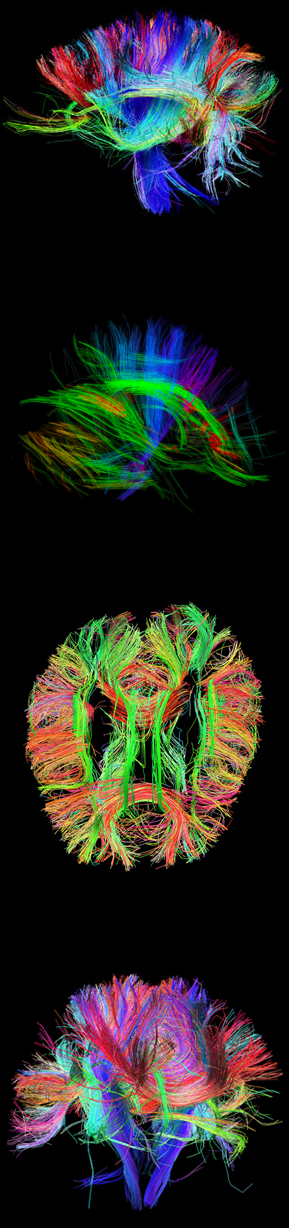
Mapping of the human connectome offers a unique opportunity to understand the complete details of neural connectivity (Sporns et al., 2005, Wedeen et al., 2008, Hagmann et al., 2007). The Human Connectome Project (HCP) is a project to construct a map of the complete structural and functional neural connections in vivo within and across individuals. The HCP represents the first large-scale attempt to collect and share data of a scope and detail sufficient to begin the process of addressing deeply fundamental questions about human connectional anatomy and variation.
Human Connectome Project Pamphlet (web-resolution)
USC-Harvard Consortium
Through a collaboration between the Laboratory of Neuro Imaging and Martinos Center for Biomedical Imaging at Massachusetts General Hospital, the HCP is being developed to employ advanced neuroimaging methods, and to construct an extensive informatics infrastructure to link these data and connectivity models to detailed phenomic and genomic data, building upon existing multidisciplinary and collaborative efforts currently underway. Working with the HCP consortium based at Washington University in St. Louis, we will provide rich data, essential imaging protocols, and sophisticated connectivity analysis tools for the neuroscience community.
The Human Connectome Project is a five-year project sponsored by sixteen components of the National Institutes of Health, split between two consortia of research institutions. Funding for the Harvard/MGH-USC consortium is provided through the grant award U01-MH93765. To read an overview of the consortia, see the NIH Blueprint Human Connectome.
Methods
The HCP is leveraging key scientific domains that together yield a steady release of increasingly detailed connectomics data and tools. First, we have begun amassing data for the release of a very large, existing connectomic, behavioral and genomic dataset, including a large sample study in MZ/DZ twin pairs, will encourage broad participation in the HCP by the larger research community. These rich data will also allow us to quantify genetic (Chiang et al., 2009) and behavioral variation of white matter fiber pathways and functional correlations for analysis by the entire community, and help define an optimized methodology for collection of a definitive connectome dataset using DSI (V. J. Wedeen, 2005). Concurrently, we are working to refine and optimize the spatial and functional resolution of our connectome neuroimaging techniques, then bring the results of both goals to bear in the acquisition of the optimized HCP data, to be shared with the community as the data are acquired. Additionally, our connectome efforts include the acquisition of high resolution neuroimaging data in a small subset of ex vivo whole brain specimens, as well as detailed chemo- and cyto-architectonic analysis and planar polarimetry of these specimens, will allow us examine correlation between cytoarchitecture and the connectome (Burgel et al., 2006), as well as help validate our in vivo results. All the while, we will continuously build and refine vital infrastructure to support the analysis, databasing and querying, and broad-scale dissemination of our data and informatics tools.
Results
This project is presently working to achieve the following goals: 1) develop sophisticated tools to process high-angular diffusion (HARDI) and diffusion spectrum imaging (DSI) from normal individuals to provide the foundation for the detailed mapping of the human connectome; 2) optimize advanced high-field imaging technologies and neurocognitive tests to map the human connectome; 3) collect connectomic, behavioral, and genotype data using optimized methods in a representative sample of normal subjects; 4) design and deploy a robust, web-based informatics infrastructure, 5) develop and disseminate data acquisition and analysis, educational, and training outreach materials.
Conclusions
Through this comprehensive white matter mapping project we will provide the neuroscience research community with a novel resource for connectomics that will have a significant impact for our enhancing our understanding of the rich neuroanatomical connectedness of the human brain.
References
BURGEL, U., AMUNTS, K., HOEMKE, L., MOHLBERG, H., GILSBACH, J. M. & ZILLES, K. (2006) White matter fiber tracts of the human brain: three-dimensional mapping at microscopic resolution, topography and intersubject variability. Neuroimage, 29, 1092-105.
CHIANG, M. C., BARYSHEVA, M., SHATTUCK, D. W., LEE, A. D., MADSEN, S. K., AVEDISSIAN, C., KLUNDER, A. D., TOGA, A. W., MCMAHON, K. L., DE ZUBICARAY, G. I., WRIGHT, M. J., SRIVASTAVA, A., BALOV, N. & THOMPSON, P. M. (2009) Genetics of brain fiber architecture and intellectual performance. J Neurosci, 29, 2212-24.
HAGMANN, P., KURANT, M., GIGANDET, X., THIRAN, P., WEDEEN, V. J., MEULI, R. & THIRAN, J.-P. (2007) Mapping Human Whole-Brain Structural Networks with Diffusion MRI. PLoS ONE, 2, e597.
SPORNS, O., TONONI, G. & KOTTER, R. (2005) The human connectome: A structural description of the human brain. PLoS Comput Biol, 1, e42.
V. J. WEDEEN, P. H., W.-Y. I. TSENG, T. G. REESE AND R. M. WEISSKOFF. (2005) Mapping complex tissue architecture with diffusion spectrum magnetic resonance imaging. . Mag. Res. Med., 54, 1377-86.
WEDEEN, V. J., WANG, R. P., SCHMAHMANN, J. D., BENNER, T., TSENG, W. Y., DAI, G., PANDYA, D. N., HAGMANN, P., D’ARCEUIL, H. & DE CRESPIGNY, A. J. (2008) Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage, 41, 1267-77.